Getting the picture - part one
/Science is mostly marketing. Sure, there's all the labor required by brainstorming new ideas and carrying out experiments, but in the end, the results have to get to their intended audience. Much as with marketing a product to potential customers, we often don't know the ideal audience for a set of scientific findings. Who will find them interesting enough to build upon them? Who will even understand them or how they may be useful?
One of my networks in its raw form. I could re-arrange it but it still wouldn't make any sense. It needs to be trimmed, or maybe even decimated.
We publish our findings. We give talks and keep asking questions. The smallest details can make the difference between our audiences understanding those results and simply ignoring them among the overwhelming volumes of information each of us swims in daily.
So, we need balance. We need smooth, logical transitions. We need to trim excess detail without losing the soul of our conclusions.
Some researchers feel that open data, in its purest form, is the best solution. It can't hurt. I'm not just talking about data here, though: I'm concerned with interpretation. I suspect that effective communication of scientific findings is a more deeply-rooted and philosophical issue than data access ever has been. Science is intended to be complex! It concerns complex issues, so our interpretation (and our visual interpretation, especially) will be unavoidably complex.
It can't stay that way.
Here are a few visualization tools I've found lately. I won't discuss them exhaustively, but in the context of balanced, efficient communication. I'll continue to post about such tools as I find them.
A recently-made slide on Slides.com. Not the best example, perhaps, but it's a start.
I'm fond of Google Slides, though mostly because it's cloud-based, a feature which aids cross-platform compatibility. It's nice to be able to give a presentation virtually anywhere and remain confident it will perform the same way. The same is not true for Powerpoint. Slides.com adds a few unique features: the ability to control a presentation with a mobile device (Google Slides requires a Chromecast for this option, I think), SVG support, and full HTML edit-ability (plus custom CSS, though that's a paid option).
Not every talk has to be a TED talk (in fact, I'd argue that most talks shouldn't be) but they should be a series of atomic visual concepts built upon each other.
Update: I tried out Slides.com - it's easy to use and the results are quite attractive. It has a few notable downsides: all presentations made with free accounts are public, creating tables is painful as cut and paste isn't an option, and visual customization isn't as simple as it ought to be (if you're used to the Slide Master in Powerpoint or even Google Slides, you may be disappointed). On the positive side, there are some unique web-centric options like embedded iFrames, plus presentations are non-linear so you can provide the appropriate visual aid if an audience member elects to interrupt you with probing questions.
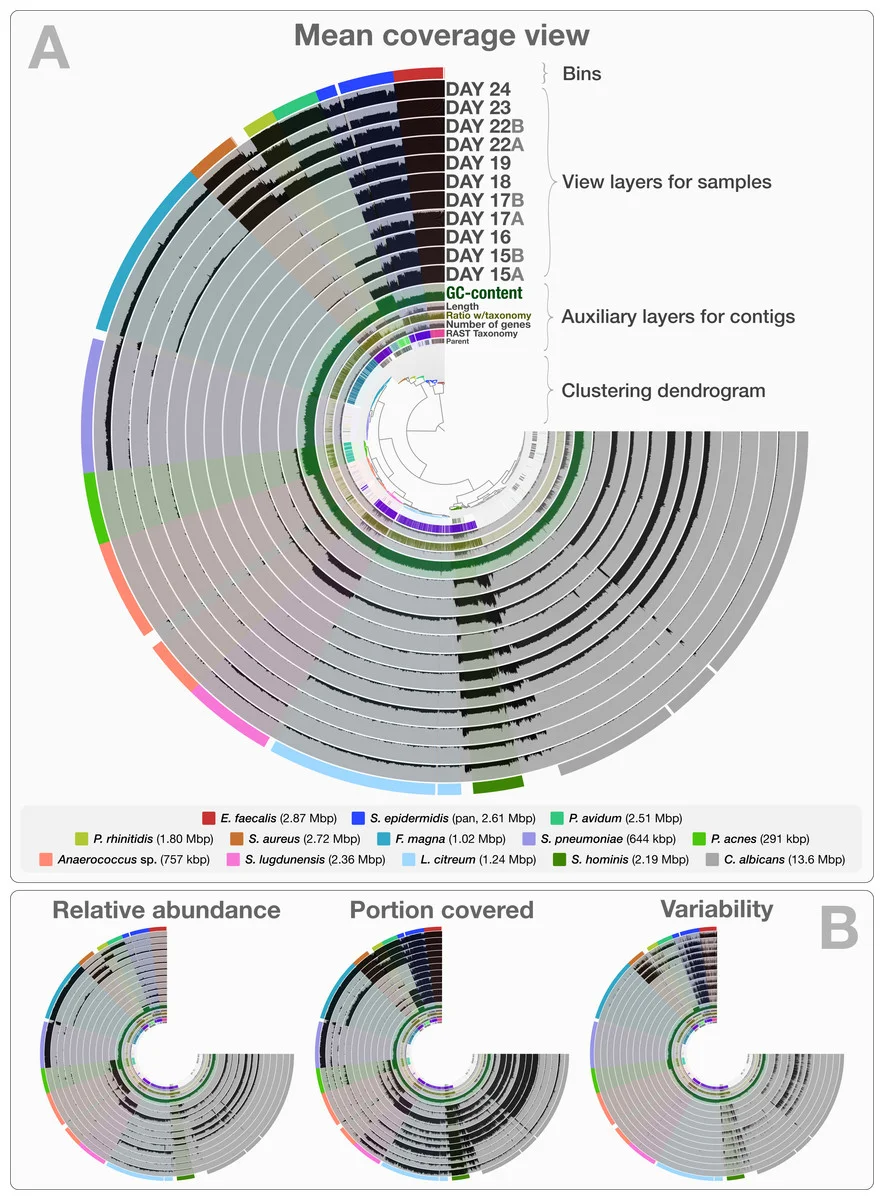
Fig. 2 from Eren et al. (2015). You spin me right round, baby, right round.
Intended for visualizing metagenomic sequence sets, anvi'o aims to be comprehensive yet easily usable, especially for researchers who actually want to observe differences between isolates. The output reminds me of Circos. It's notable here because, with metagenome or pangenome data sets, the biggest conclusion is often that we have the data in the first place. Drilling down to the sequence level is even better (though, as usual, I'm curious about how it handles viral genomes).
There's a short intro to using anvi'o for microbial pangenomics here, courtesy of its authors.
I haven't had a chance to play with anvi'o yet but it may be the difference between "this is a pangenome" and "we know which sequences aren't consistent across the pangenome".